 |
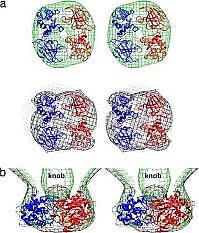
Figure: Stereo diagrams showing two gp13 monomers (blue and red) fitted into the 2-fold averaged cryoEM density of the emptied WT (green contours) and mutant full sus13(330) (black contours) cone densities. End-on (a) and side (b) views of a 40-Å-thick cross-section. |
Michael Rossmann's group has published a virology paper, using data collected at GM/CA-CAT, on X-ray crystallography and cryoEM structural studies of a bacterial-cell-wall-degrading enzyme in the bacteriophage phi29 tail. The small bacteriophage phi29 must penetrate the approximately 250 Å thick external peptidoglycan cell wall and cell membrane of the Gram-positive Bacillus subtilis, before ejecting its dsDNA genome through its tail into the bacterial cytoplasm. The tail of bacteriophage phi29 is non-contractile and approximately 380 Å long. A 1.8-Å resolution crystal structure of gene product 13 (gp13) shows that this tail protein has spatially well separated N- and C-terminal domains, whose structures resemble lysozyme-like enzymes and metallo-endopeptidases, respectively. Cryo-electron microscopy reconstructions of the wild-type bacteriophage and mutant bacteriophages missing some or most of gp13 shows that this enzyme is located at the distal end of the phi29 tail knob. This suggests that gp13 functions as a tail-associated-peptidoglycan degrading enzyme able to cleave both the polysaccharide backbone and peptide crosslinks of the peptidoglycan cell wall. Comparisons of the gp13- mutants with the phi29 mature and emptied phage structures suggest the sequence of events that occur during the penetration of the tail through the peptidoglycan layer that normally protects Gram-positive bacteria.
Citation:
Xiang, Y, Morais, MC, Cohen, DN, Bowman, VD, Anderson, DL, Rossmann, MG.
Crystal and cryoEM structural studies of a cell wall degrading enzyme in the
bacteriophage {phi}29 tail, Proc. Natl. Acad. Sci. USA 105 (28), 9552-9557
(2008). DOI: 10.1073/pnas.0803787105.