The group of Michael Malkowski at the Hauptman-Woodward Medical Research Institute and Department of Structural Biology, SUNY at Buffalo, determined the x-ray crystal structure of Arabidopsis thaliana alpha-dioxygenase (alpha-DOX) to 1.5-Å resolution using MAD phasing, which yielded insight into diversity with the cyclooxygenase-peroxidase superfamily of monotopic membrane enzymes. alpha-DOX is a heme-containing enzyme that catalyzes the oxygenation of fatty acids into 2(R)-hydroperoxide products via the stereospecific removal of the pro-R hydrogen from carbon-2 of the fatty acid substrate. Despite the low level of sequence identity, alpha-DOX shares common catalytic features with cyclooxygenase (COX) within the cyclooxygenase-peroxidase superfamily, including the use of a tyrosyl radical during catalysis. The overall structure of alpha-DOX is predominantly alpha-helical in nature and comprised of a base domain and a catalytic domain. The base domain exhibits a low degree of structural homology with the membrane-binding domain of COX. The catalytic domain shows the highest degree of similarity with COX, where 21 of the 22 alpha-helical elements are conserved. Four alpha-helices form the heme-binding cleft and walls of the active site channel, which are highly similar to the alpha-helices that define the cyclooxygenase channel in COX. These four alpha-helices contain the proximal and distal histidines required for heme binding, the catalytic tyrosine, as well as highly conserved arginine and histidine residues involved in fatty acid binding and positioning. Unlike COX, the heme-binding cleft is buried deep within the catalytic domain of the enzyme, where access to the distal face of the heme is restricted by the presence of two extended inserts unique to alpha-DOX. The alpha-DOX structure represents the first member of the alpha-DOX subfamily to be structurally characterized within the cyclooxygenase-peroxidase family of heme-containing proteins.
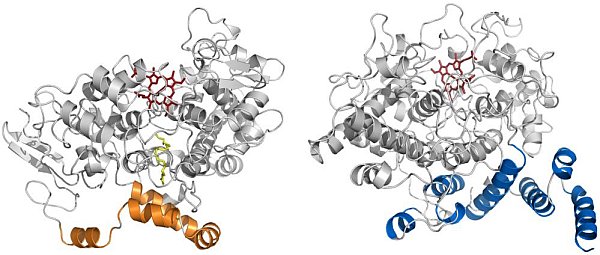 |
Figure: Comparison of the structures of COX and alpha-DOX. (Left) Ribbon diagram of mouse COX-2 (PDB id 3HS5) with arachidonic acid (yellow) bound within the cyclooxygenase channel. The alpha-helices of the membrane-binding domain are colored in gold. (Right) Ribbon diagram of alpha-DOX. The alpha-helices of the base domain are colored in blue. The heme moiety in each structure is colored red. |
Citation:
Goulah, CC, Zhu, G, Koszelak-Rosenblum, M, Malkowski, MG. The Crystal
Structure of [alpha]-Dioxygenase Provides Insight into Diversity in the
Cyclooxygenase-Peroxidase Superfamily, Biochemistry 52, 1364-1372 (2013).
DOI: 10.1021/bi400013k