The groups of Amie Boal and Squire Booker at the Pennsylvania State University determined the structure of the dual-specificity RNA methylase, RlmN, cross-linked to a tRNA substrate. RlmN, which is a member of the radical S-adenosylmethionine (SAM) superfamily of enzymes, methylates both ribosomal and transfer RNAs at the C2 position of specific adenine nucleotides using a unique radical mechanism that involves a cross-linked protein/nucleic acid intermediate. The methylation of A2503 in domain V of 23S rRNA has been reported to enhance translational fidelity. By changing a key cysteine residue to alanine, the groups were able to trap an RlmN-tRNA cross-linked structure at this intermediate step and solve its structure, giving insight into substrate recognition and several unresolved details of the mechanism. The structure is the first view of a dual-specificity methylase bound to a full-length RNA substrate. RlmN forms an extended interface with tRNA, which is rare among tRNA-modifying enzymes, but is similar to the binding mode of tRNA synthetases; this provides an unusual induced-fit mechanism for binding the tRNA, which completely disrupts the structure of the anticodon stem-loop. The capacity to significantly remodel the substrate is likely key in the ability of RlmN to target two different forms of RNA, and is distinct from the only other known dual-specificity RNA modifying enzyme, RluA. RlmN has a close evolutionary relationship and enzymatic mechanism to Cfr, which methylates the C8 position of the same nucleotide (A2503) in the same rRNA. In contrast to RlmN, Cfr confers resistance to several different classes of clinically-used antibiotics. The structure of RlmN in complex with an RNA substrate could aid in structure-based design of new antibacterial agents targeting Cfr.
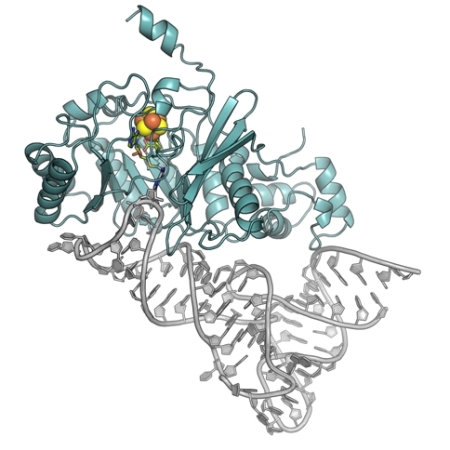 |
Figure: Cartoon diagram of RNA-modifying enzyme RlmN (blue) bound to tRNA (gray). The space-filling iron-sulfur cluster is colored by atom type. The cross-link and SAM cleavage products are shown in stick format and are colored by atom type. |
Citation: Schwalm, E.L., Grove, T.L., Booker, S.J., and Boal, A.K. (2016) Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science 352, 309-312.