The group of Martin Caffrey determined the structure of lipoprotein signal peptidase II (LspA) complexed with globomycin, a promising candidate in the fight to overcome antibiotic resistance. LspA is a bacterial integral-membrane aspartyl peptidase that processes lipoproteins, which play critical roles in bacterial physiology, pathogenicity, and antibiotic resistance. LspA cleaves the signal peptide that anchors the lipoprotein to the cytoplasmic membrane, with the extrinsic domain in the periplasm or outside the cell. LspA is essential in many pathogenic bacteria, but there is no equivalent in humans, making it a potential drug target. Globomycin is a natural antimicrobial produced by select strains of Streptomyces. The cyclic depsipeptide is effective against Gram-negative bacteria, and synthetic analogs also inhibit the growth of Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA). Growing resistance to previously-effective antibiotics is a major concern around the world. Indeed, this world challenge required an international effort, where the team led by Caffrey brought their LCP-grown crystals to GM/CA@APS beamlines, as well as to beamlines at the Swiss Light Source and at Diamond, to reveal the structure of the complex. Vogeley et al. determined the crystal structure of LspA from Pseudomonas aeruginosa, an opportunistic human pathogen, complexed with the antimicrobial at 2.8 Å resolution. Globomycin acts as a non-cleavable peptide that blocks the LspA active site. Conserved residues are networked to such a degree around the active site that mutations interfering with antibiotic binding would likely also impair substrate binding or catalysis. These findings should be useful in the development of new antibiotics.
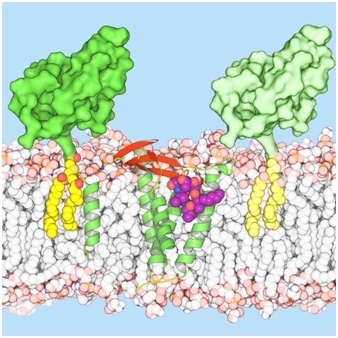 |
Figure: LspA bound to globomycin in the center of the membrane bilayer (LspA shown with green α-helices and red β-strands; globomycin as purple, orange, and blue spheres). Lipoproteins (bulky green), anchored to the membrane by lipids (yellow) are shown to either side, where the lipoprotein on the left also has a hydrophobic signal peptide in the form of a green helix. |
Citation: Vogeley, L., El Arnaout, T., Bailey, J., Stansfeld, P.J., Boland, C., and Caffrey, M. (2016) Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 351, 876-880.