Using beamline 23ID-D, the group of Guillermo Calero at the University of Pittsburgh and collaborators determined the structure of human uracil DNA glycosylase (UNG2) in complex with an HIV protein and with two other human proteins that are involved in cellular degradation. UNG2 is a DNA repair enzyme whose role in the development of AIDS has been highly controversial. Base excision repair (BER) is the cellular process where non-deforming lesions are removed from DNA. Such lesions are highly deleterious to the cellular genome. The first stage of BER requires DNA glycosylases, which recognize and remove damaged or "mispaired" bases, leaving an abasic (AP) site. It has been estimated that over 10,000 AP sites are generated in a cell per day. During later BER stages, large molecular machines remove and replace AP sites in a lengthy process that leads to full repair of the original lesion. UNG2 serves to remove uracil lesions from DNA, leaving an AP site. A controversy has existed regarding its role in the development of AIDS, in that reports indicate that UNG2 can have "favorable effects" for the HIV-1 virus, since it can help maintain viral RNA genome stability, as well as "unfavorable effects" for HIV-1, since it would damage the HIV genome by creating AP sites. The crystal structure of UNG2 in complex with the accessory HIV Viral protein R (Vpr) and two other human proteins determined in this study sheds light on this controversy. It illustrates the elegant mechanism employed by Vpr to "mimic" damaged-DNA, thus binding to UNG2 and inhibiting its activity, and therefore impairing repair of the heavily uracilated HIV genome. However, inhibition is not enough: the complex also contains human damage-specific DNA-binding protein 1 (DDB1), and human DDB1-CUL4-associated factor 1 (DCAF1), which is a substrate receptor for the Cullin4-RING finger E3 ubiquitin ligase (CRL4) of the host ubiquitin proteasome-mediated protein degradation system. This structure of the multi-protein complex reveals that in addition to inhibition, Vpr steers UNG2 towards destruction, using the host degradation machinery, and thereby knocking out UNG2 with a two-step punch. International interest in developing ways to combat HIV-1, and the recent focus on the interplay between virus and the host, make these findings extremely relevant at this time.
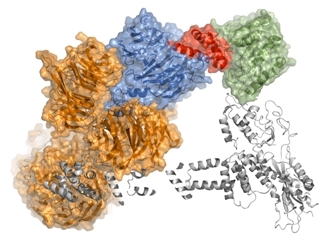 |
The figure shows the crystal structure of DDB1 (orange), DCAF1 (marine), Vpr (red), and UNG2 (green), as well as a model of potential interactions with CRL4 (grey ribbons). |
Wu Y, Zhou X, Barnes CO, DeLucia M, Cohen AE, Gronenborn AM, Ahn J, Calero G, "The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction," Nat. Struct. Mol. Biol. 23, 933-940 (2016). DOI: 10.1038/nsmb.3284