In both eukaryotes and prokaryotes, protein secretion across membranes is a fundamental cellular process, whether co-translational during protein synthesis, or post-translational during secretion. In the general secretion pathway, the Sec translocon transports many secreted and plasma membrane proteins across the eukaryotic endoplasmic reticulum or across bacterial plasma membranes. In bacteria, a transmembrane protein, SecY, is central to both processes. For post-translational secretion, it forms a complex with water-soluble SecA, which uses ATP to actively transport peptides across the plasma membrane. Previously, the group of Tom Rapoport at Harvard University had reported crystal structures of the idle channel, and of a complex of the channel with the translocation ATPase SecA, but a detailed structure of an inserting polypeptide in an active channel had been elusive. Using beamline 23ID-D (during last year's CCP4/APS crystallography school hosted by GM/CA@APS), Long Li in the Rapoport group and collaborators determined a crystal structure of the active protein translocation channel, which had been a "holy grail" in the field. The new structure contains the SecY channel, SecA, and a short segment of a secretory protein, inserted into the channel as a loop. The hydrophobic region of the signal sequence forms a helix that sits in a groove outside a lateral gate of the SecY channel. The C-terminal section of the polypeptide loop is located in the channel. The structure shows that the hydrophobic regions of signal sequences exit the lateral gate and partition into the lipid phase, explaining their diversity in sequence and length. A comparison with other structures shows that this mechanism of signal-sequence recognition is universal, regardless of the organism and mode of translocation, and also applies to bilayer-spanning domains of nascent membrane proteins.
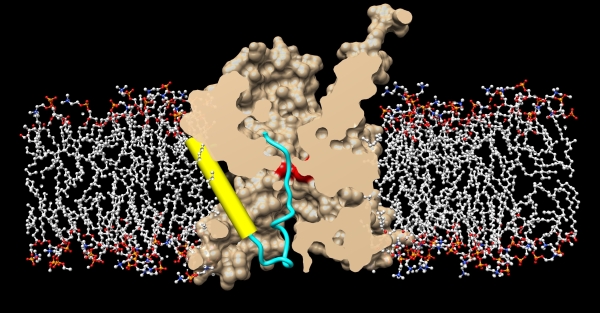 |
Figure: Crystal structure of the active protein translocation channel, where SecY is shown in beige, and the membrane bilayer is shown in stick models. The SecY pore ring is shown in red, the translocating polypeptide is show in blue, and the hydrophobic signal sequence is shown in yellow, exiting through the lateral gate. |
Citation: Li, L., Park, E., Ling, J.J., Ingrame, J., Ploegh, H. and Rapoport, T.A. (2016) Crystal structure of a substrate-engaged SecY protein-translocation channel, Nature 531, 395-399.