Janet Smith's group from the University of Michigan used the beamline automounter to screen ~48 crystals in ~4 hrs to solve the structure of the ECH2 domain of CurF from Lyngbya majuscula. CurF ECH2 is a decarboxylase involved in the introduction of a cyclopropyl ring in the biosynthesis of the natural product curacin A, which is a potent cytotoxic agent. The structure was solved with a 3-wavelength MAD data set using selenomethionine-labeled protein on beamline 23ID-B. The structure demonstrated that CurF ECH2 has a crotonase fold, and substrate modeling into the active site facilitated testing of active site residues. This new structure has provided insights into β-branching in polyketide synthesis as described in a manuscript submitted for publication.
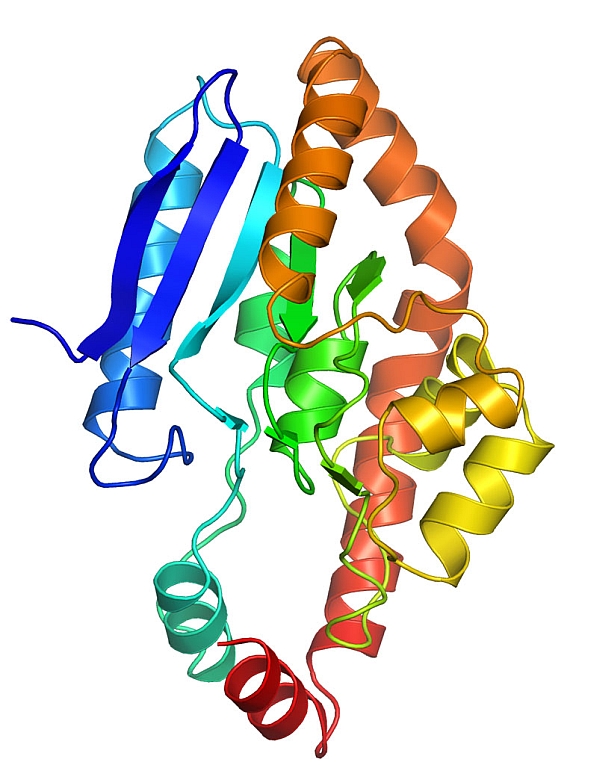 |
Figure: View of CurF ECH2 monomer demonstrating crotonase superfamily fold and active site chamber highlighted with red asterisk. |
Citation:
Geders, TW, Gu, L, Mowers, JC, Liu, H, Gerwick, WH, Hakansson, K, Sherman,
DH, Smith, JL. "Crystal Structure of the ECH2 Catalytic Domain of
CurF from Lyngbya majuscula: Insights into a Decarboxylase Involved in
Polyketide Chain β-Branching", J. Biol. Chem. 282 (49), 35954-35963
(2007). DOI: 10.1074/jbc.M703921200.