Peter Kuhn's group from The Scripps Research Institute has provided several examples of structures determined from the SARS-CoV proteomics project using data from GM/CA-CAT (as well as from SSRL) . These three structures include two important non-structural proteins, nsp10 and nsp15, and the third was an RNA-binding domain of the SARS nucleocapsid. The 2.9-Å Nsp15 structure of a deletion construct truncated at both the C- and N- terminus required extensive screening of multiple protein constructs. This structure revealed that truncation of the N-terminal oligomerization domains turns it from a hexameric endonuclease into an obligate monomer which is inactive. The primary reason for the loss of catalytic activity was found to be a dramatic collapse of the two active site loops that support the nucleotide ligand and metal ion during catalysis.
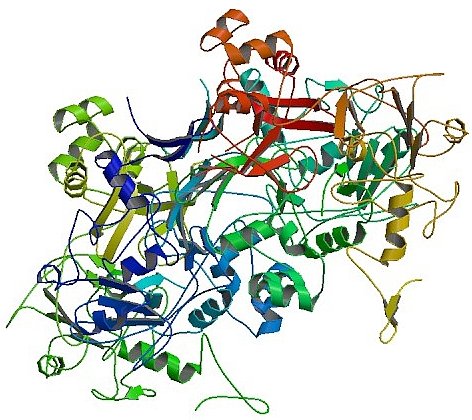 |
Figure: Structure of an N-Terminal Truncated Form of Nendou (NSP15) From SARS-CORONAVIRUS [PDB ID 2OZK] |
Citation:
| [1] | Joseph, JS, Saikatendu, KS, Subramanian, V, Neuman, BW, Brooun, A, Griffith, M, Moy, K, Yadav, MK, Velasquez, J, Buchmeier, MJ, Stevens, RC, Kuhn, P. Crystal Structure of Nonstructural Protein 10 from the Severe Acute Respiratory Syndrome Coronavirus Reveals a Novel Fold with Two Zinc-Binding Motifs, J. Virol. 80 (16), 7894-7901 (2006). DOI: 10.1128/JVI.00467-06. |
| [2] | Joseph, JS, Saikatendu, KS, Subramanian, V, Neuman, BW, Buchmeier, MJ, Stevens, RC, Kuhn, P. Crystal Structure of a Monomeric Form of Severe Acute Respiratory Syndrome Coronavirus Endonuclease nsp15 Suggests a Role for Hexamerization as an Allosteric Switch, J. Virol. 81 (12), 6700-6708 (2007). DOI: 10.1128/JVI.02817-06. |