The group of Harry Noller at the University of California at Santa Cruz extended their previous studies on translation termination. Bacterial translation termination depends on release factors, RF1 and RF2, which recognize stop codons and hydrolyze the peptidyl-tRNA ester bond. The structure of a translation-termination complex with RF2 followed the group's publication of an RF1 complex last year. The specificity for stop-codon recognition by both RF1 (UAG and UAA) and RF2 (UGA and UAA) occurs in structural motifs of domain 2. In both structures rearrangement of a switch loop places a conserved GGQ motif at the catalytic site of peptidyl-tRNA hydrolysis. Biochemical experiments support an unusual mechanism in which the main-chain amide nitrogen of the conserved glutamine in the GGQ motif participates directly in catalysis.
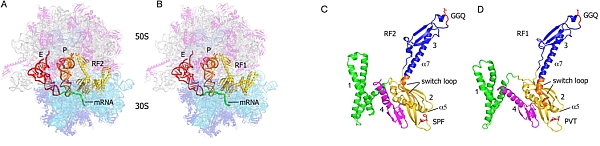 |
Figure: RF1 and RF2 termination complexes. (A) RF2 termination complex showing RF2 (yellow), P-site tRNA (orange), E-site tRNA (red), mRNA (green), 16S rRNA (cyan), 23S and 5S rRNA (gray), 30S proteins (blue), and 50S proteins (magenta). (B) RF1 termination complex colored similarly to A. (C) RF2 in the ribosome-bound conformation. (D) RF1 in the ribosome-bound conformation. |
Citation:
Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Zhu, J,
Trakhanov, S, Scott, WG, Noller, HF, Crystal structure of a translation
termination complex formed with release factor RF2, Proc. Natl. Acad. Sci.
USA 105 (50), 19684-19689 (2008). DOI: 10.1073/pnas.0810953105