Mika Jormakka's group of the University of New South Wales (and now of the University of Sydney) in Australia has determined the structure of a membrane protein fundamental to polysulfide respiration in bacteria in extreme environments. The enzyme uses quinones to pump protons across the membrane. The structure of polysulfide reductase (Psr) from Thermus thermophilus was determined to 2.4-Å resolution. Psr has a homo-dimer configuration, composed of two membrane-associated subunits (A and B), and one integral-membrane subunit (C). Quinone-inhibitor-bound structures revealed an electron-transfer pathway from the membrane domain to the active site situated in the A subunit, including a water-filled cavity connecting the quinone binding site on the periplasmic side to the cytoplasmic side. Structured water molecules in the membrane domain indicate that Psr is capable of proton pumping through putative long-range conformational changes. The structures collectively provide seminal insights into the mechanism of energy conservation.
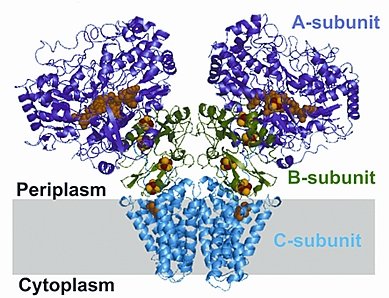 |
Figure: Psr has two membrane-associated subunits (A and B), and one integral-membrane subunit (C). |
Citation:
Jormakka, M, Yokoyama, K, Yano, T, Tamakoshi, M, Akimoto, S, Shimamura, T,
Curmi, P, Iwata, S. Molecular mechanism of energy conservation in
polysulfide respiration, Nat. Struct. Mol. Biol. 15, 730-737 (2008). DOI:
10.1038/nsmb.1434.