Michael Rossmann's group at Purdue University employed a combination of X-ray crystallography and electron cryo-microscopy (cryo-EM) to investigate DNA packaging into procapsids. Sun et al. solved the structure of gene product 17 (gp17), and fit it to the EM map. Pentameric gp17 associates with a unique vertex of the procapsid head. Fitting the crystal structure of gp17 into the EM map showed a more extended arrangement of the N- and C-terminal domains of gp 17 in the EM map than in the crystal structure. Based on these "relaxed" and "tense" states of gp17, a multi-step model of DNA translocation into the procapsid was proposed.
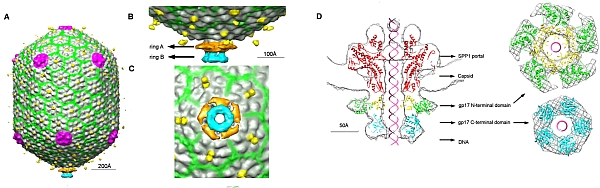 |
Figure: (A) Cryo-EM map of the T4 procapsid with bound gp17 showing viral capsid proteins in gray, pink, yellow and green, and the gp17 N-terminal domain in orange and C-terminal domain in cyan. (B) Side view of the portal with gp17. (C) Bottom view of the portal showing the pentameric gp17. (D) The dodecameric portal protein SPP1 (red) and gp17 N-terminal subdomain I (green), subdomain II (yellow) and C-terminal domain (cyan) fit to the cryo-EM density. |
Citation:
Sun, S, Kondabagil, K, Draper, B, Alam, TI, Bowman, VD, Zhang, Z, Hegde, S,
Fokine, A, Rossmann, MG, Rao, VB. The Structure of the Phage T4 DNA
Packaging Motor Suggests a Mechanism Dependent on Electrostatic Forces, Cell
135 (7), 1251-1262 (2008). DOI: 10.1016/j.cell.2008.11.015