The Structural Genomics Consortium at the University of Toronto solved the structure of the SRA domain of human UHRF1, which recognizes hemi-methylated DNA. DNA methylation of CpG dinucleotides is an important epigenetic modification of mammalian genomes that is critical for regulation of genome expression, genome stability, and chromatin structure; changes in DNA methylation patterns are important in development, and aberrant changes may be related to carcinogenesis. UHRF1 is an E3 ligase that is important for the maintenance of DNA methylation in vivo, in that its SET and Ring-associated (SRA) domain recognizes hemi-methylated DNA and that it associates with maintenance DNA methyl transferase I (DNMT1). The structures of the apo SRA domain of human UHRF1 and of the SRA domain complexed with hemi-methylated DNA show how 5-methylcytosine flips out of a DNA duplex to enter the SRA domain, and how two SRA loops reach into the gap in the DNA duplex to read the other three bases of the CpG duplex. The major-groove loop provides specificity for binding of the CpG dinucleotide, and discrimination against methylation of deoxycytidine of the complementary strand, a key factor for DNMT1 maintenance methylation.
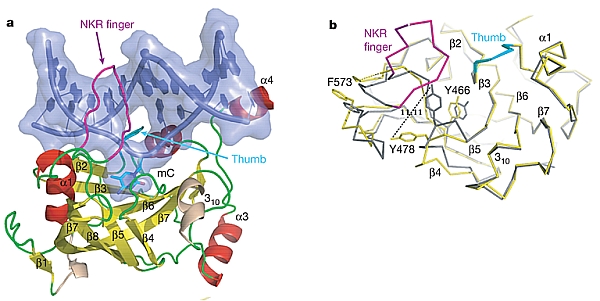 |
Figure: (A) DNA-SRA complex. The flipped, methylated deoxycytidine (mC); the resulting hole in the DNA is partially filled by the NKR finger (magenta) and the thumb (cyan). (B) Superposition of the apo (yellow) and DNA-bound (grey) SRA. [ Figure reprinted by permission from Macmillan Publishers Ltd: Nature, 2008] |
Citation:
Avvakumov, GV, Walker, JR, Xue, S, Li, Y, Duan, S, Bronner, C, Arrowsmith,
CH, Dhe-Paganon, S. Structural basis for recognition of hemi-methylated DNA
by the SRA domain of human UHRF1, Nature 455, 822-826 (2008). DOI:
10.1038/nature07273