The group of Sylvie Doublié at the University of Vermont solved structures of two 8-oxoguanine DNA glycosylases, which recognize specific DNA lesions and initiate repair processes. Guanine is the most vulnerable of DNA bases to oxidative damage; its most common oxidative product, 7,8-dihydro-8-oxoguanine (8-oxoG), is the most prevalent defect found in DNA molecules. In the base-excision DNA repair pathway, 8-oxoG is recognized by 8-oxoguanine DNA glycoslyase (Ogg), which cleaves the N-glycosylic bond and then the DNA backbone. Ogg enzymes occur in three families: Ogg1, Ogg2, and AGOG. This work provides the first Ogg2 structures, from Methanocaldococcus janischii (MjaOgg) and Sulfolobus solfataricus (SsoOgg). The structures show that the C-terminal lysine of Ogg2 plays a key role in discriminating between guanine and 8-oxoG in Ogg2. Comparison with the structure of human Ogg1 shows some conserved features in the C-terminal region for binding 8-oxoG, as well as some key differences.
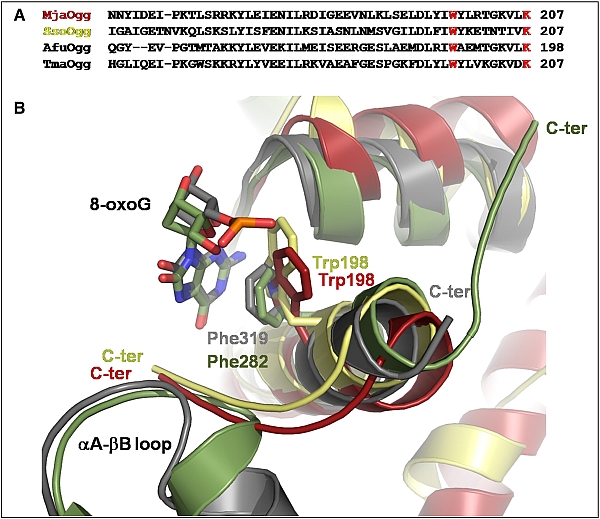 |
Figure: (A) Alignment of the C-terminal region of several Ogg2 sequences, showing the conserved "stacking" tryptophan and the C-terminal lysine (red). (B) Ribbon diagram showing a superposition of the C-terminal loop of MjaOgg (red), SsoOgg (yellow), the ?A-?B loop of human Ogg1 (gray) (PDB 1EBM), and CacOgg (green) (PDB 3F10). The superposition shows the importance of an aromatic stacking interaction (Trp198 in Ogg2) with the 8-oxoG substrate. |
Citation:
Faucher, F, Duclos, S, Bandaru, V, Wallace, SS Doublié, S. Crystal
Structures of Two Archaeal 8-Oxoguanine DNA Glycosylases Provide Structural
Insight into Guanine/8-Oxoguanine Distinction, Structure 17 (5), 703-712
(2009). DOI: 10.1016/j.str.2009.03.007