Cynthia Wolberger's group at the Johns Hopkins University determined the structure of the deubiquinating module (DUBm) of the 1.8-MDa SAGA complex that regulates covalent modifications of chromatin. DUBm is a four-protein component of SAGA whose deubiquinating function helps remove histones from the promoter regions of genes. DUBm is composed of Ubp8, a ubiquitin-specific protease, and the Sgf11, Sus1, and Sgf73 subunits. In structures of apo-DUBm and the ubiquitin aldehyde complex, all four subunits are strongly intertwined, and the module is stabilized by eight Zn atoms that are critical for enzymatic activity. Seven Ubp8 residues near the active site are disordered in the absence of ubiquitin. The structures led to plausible models for Sgf11/Sus1/Sgf73 activation of Ubp8 and for DUBm recognition of monoubiquinated histone H2B.
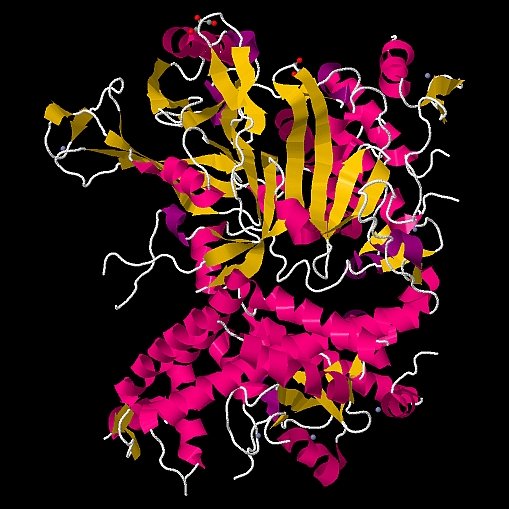 |
Figure: the 1.8-MDa SAGA complex [ PDB ID 3MHS ] |
Citation:
Samara, NL, Datta, AB, Berndsen, CE, Zhang, X, Yao, T, Cohen, RE, Wolberger,
C, Structural Insights into the Assembly and Function of the SAGA
Deubiquitinating Module, Science 328, 1025-1029 (2010). DOI:
10.1126/science.1190049