Antibiotic drug resistance is a growing global public health problem. In the bacterium Staphylococcus aureus, the enzyme Cfr confers resistance to antibiotics by methylating the C8 position of adenosine 2503 (A2503), which is ultimately found in the peptidyl transferase center of the 50S subunit of the bacterial ribosome. Cfr is closely related to RlmN, which modifies the C2 position on A2503 for a housekeeping function unrelated to drug resistance. These two enzymes use Sadenosylmethionine (SAM) as both the source of the methyl group and the source of a 5'-deoxyadenosyl 5'-radical (5'-dA·) generated using a [4Fe-4S] cluster. In the proposed mechanism, the enzyme first appends a methyl group to a conserved cysteine in the active site (Cys 355 in RlmN) in a standard SAM-dependent methyl transfer reaction. A second molecule of SAM is then reductively cleaved to activate the Cys-appended methyl group which is transferred to the appropriate carbon atom on A2503. Crystal structures of RlmN determined by researchers in the labs of Amy Rosenzweig and Squire Booker (Pennsylvania State University) provide molecular-level insight into this unusual mechanism. In the 2.05-Å resolution structure of Escherichia coli RlmN with bound SAM, the SAM cosubstrate is coordinated to a [4Fe-4S] cluster located within an active site architecture typical of radical SAM superfamily enzymes. The structure confirms the presence of a methyl substituent on Cys355, well poised for a subsequent hydrogen atom abstraction reaction to activate the methyl group. The active site arrangement also suggests an identical SAM binding site for the initial methyl transfer to Cys355, a reaction not usually catalyzed by SAM bound to an iron-sulfur cluster. Cys355 lies within a flexible loop that dips into the active site, providing a structural rationale for its multifunctional role in the reaction. A large positively charged opening to the active site provides an obvious approach for a bulky ribosomal RNA substrate. Finally, the RlmN structures help in understanding how Cfr might have evolved to facilitate antibiotic resistance.
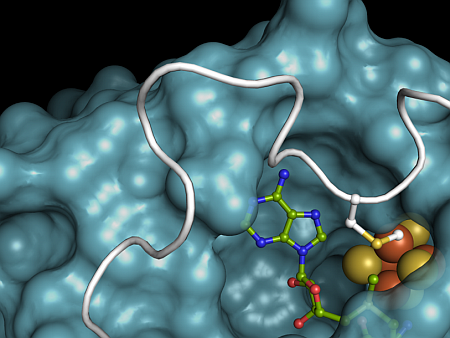 |
Figure: The RlmN active site. The [4Fe-4S] cluster is shown as orange and yellow spheres and the SAM cosubstrate is shown as sticks. Methylated residue Cys 355, shown as sticks, derives from a flexible loop region shown in white. The rest of the protein is shown as a blue surface. |
Citation: Boal AK, Grove TL, McLaughlin MI, Yennawar NH, Booker SJ, Rosenzweig AC. Structural basis for methyl transfer by a radical SAM enzyme. Science 2011 May 27; 332: 1089-92. doi: 10.1126/science.1205358.