 |
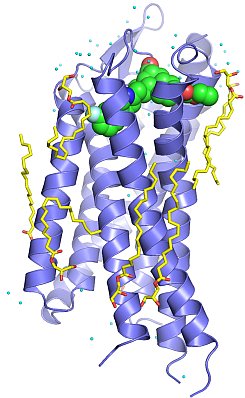
Figure: Structure of human PAR1 (blue) bound to the antagonist drug vorapaxar (green), with lipid (yellow) and water (cyan) molecules. |
Brian Kobilka's group published the first structure of a protease-activated G protein-coupled receptor (GPCR). Protease-activated receptor 1 (PAR1) is the prototypical member of a family of GPCRs that are activated by proteases. The coagulation protease thrombin activates PAR1 by specific cleavage of the receptor's N-terminal exodomain to generate a new N-terminus. This new N-terminus then functions as a tethered peptide agonist, binding intramolecularly to the seven-transmembrane bundle of the receptor to effect G protein activation. Activation of PAR1 by thrombin leads to platelet aggregation and thrombosis, and it has been considered as a promising target for developing anti-platelet drugs for treating various cardiovascular diseases. By using the T4 lysozyme insertion strategy, the Kobilka group in collaboration with Shaun Coughlin's group at UCSF solved the crystal structure of human PAR1 bound to an antagonist vorapaxar to 2.2-Å resolution. Vorapaxar is a promising drug candidate that has been evaluated in two Phase 3 clinical trials. The structure reveals an unusual ligand-binding mode of vorapaxar that explains how a small molecule binds virtually irreversibly to inhibit receptor activation by the tethered agonist peptide of PAR1. This structure will aid the development of improved PAR1 antagonists and the discovery of antagonists to other members of the protease-activated receptor family.
Citation:
Zhang, C, Srinivasan, Y, Arlow, DH, Fung, JJ, Palmer, D, Zheng, Y, Green, HF,
Pandey, A, Dror, RO, Shaw, DE, Weis, WI, Coughlin, SR, Kobilka, BK.
High-resolution crystal structure of human protease-activated receptor 1,
Nature 492, 387-392 (2012).