David Mueller's group at the Rosalind Franklin University of Medicine and Science provided insight into a key integral-membrane step in proton translocation for ATP synthesis. Mitochondrial ATP synthase is a molecular motor driven by a proton gradient. The membrane portion of the enzyme, Fo, is analogous to a proton turbine that spins within the inner mitochondrial membrane driven by the proton potential. An oligomer of 10 c-subunits forms a ring, which is the core of the proton turbine. Within subunit-c, an essential side chain carboxyl group with a pKa of 7.0 (Glu59 in the yeast enzyme), acts as the proton donor and acceptor in the rotation cycle. This study captured the yeast mitochondrial c10-ring in the "open" conformation in which the essential carboxyl group is extended and available for both protonation and deprotonation. High-resolution (2.0 Å) structures at pH 8.3 (Figure 14) and pH 5.5 correspond to the deprotonated and protonated forms, respectively. Molecular simulation studies suggest that the "open" conformation is favored when the carboxyl of Glu59 is in a more hydrophilic environment, while the "closed" state is favored in a hydrophobic environment. These states are formed after rotation of the c-ring places the carboxyl of Glu59 within the center of the lipid bilayer and alternately, within the proton access channel formed by subunit a. This study provides a critical advance in the understanding of the structural basis for proton translocation during ATP synthesis.
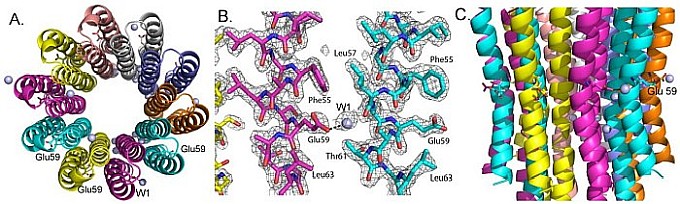 |
Figure: Yeast mitochondrial ATP synthase c10i-ring at pH 8.3. (A) The proton turbine is composed of an oligomer of 10 c-subunits arranged in a ring, here viewed from the matrix. (B) Representative electron density illustrates the structure quality. The 2.0-Å refined model, including 85 waters, had an overall B-factor of 23.2 Å2, a remarkably low value for an integral membrane protein. (C) A side view from within the mitochondrial membrane shows the Glu59 carboxyl groups involved in proton transfer, which are identical in all subunits but here labeled in just a few of the subunits. |
Citation:
Symersky, J, Pagadala, V, Osowski, D, Krah, A, Meier, T, Faraldo-Gomez, JD,
Mueller, DM. Structure of the c(10) ring of the yeast mitochondrial ATP
synthase in the open conformation, Nat Struct Mol Biol 19, 485-491 (2012).