Postdoctoral researcher, Megan Sjodt, in the laboratory of Andrew Kruse at Harvard Medical School, determined the structure of a bacterial cell wall polymerase, RodA, revealing a highly conserved, water-filled central cavity that could potentially pave the way for next-generation broad-spectrum antibacterial therapies. The structure was phased using a novel evolutionary coupling analysis. RodA is a member of the SEDS (shape, elongation, division, sporulation) family of highly conserved essential transmembrane proteins that are found in nearly all bacteria. Their function was long enigmatic, until recently when they were found to catalyze the first step in building the protective bacterial cell wall. The structure of RodA provides the first molecular insight into how disruption of RodA function causes bacterial cells to swell and die. This suggests that RodA is a viable target for the development of small-molecule inhibitors that could be effective against a wide range of bacterial pathogens.
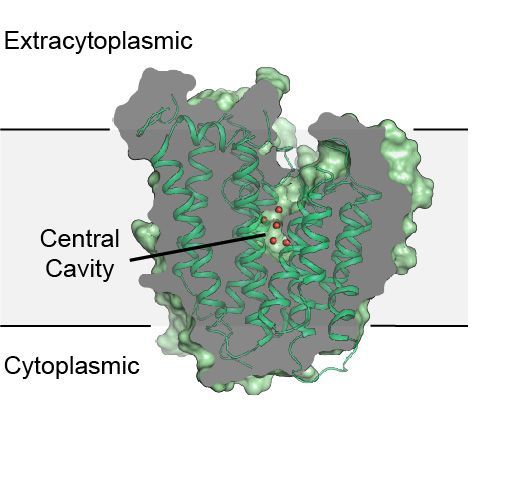 |
Figure: Structure of the transmembrane protein, RodA. |
Citation: Sjodt, M, Brock, K, Dobihal, G, Rohs, PDA, Green, AG, Hopf, TA, Meeske, AJ, Srisuknimit, V, Kahne, D, Walker, S, Marks, DS, Bernhardt, TG, Rudner, DZ, Kruse, AC. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis, Nature 556, 118-121 (2018). DOI: 10.1038/nature25985.