Engin Özkan's laboratory at the University of Chicago determined crystal structures of a group of proteins that specify the connectivity maps of neurons. These matchmaker molecules reside on neuronal cell surfaces to act as specialized glue and establish patterns of brain wiring necessary for a functional nervous system. Named Dprs and DIPs in fruit flies, they are found in all animal nervous systems. Dprs and DIPs have been reported to determine whether two neurons will make a synapse, and also to control bundling of neurons that sense smell as they enter the brain, among others. The work from the Özkan group, in collaboration with Robert Carrillo's group, reports four new three-dimensional structures of Dprs-DIPs pairs, as they would bring together two neuronal cells. The structures identify the molecular determinants of specificity within these large protein families. Using this information, the study reports the engineering of mutant fruit flies with modified synaptic connectivity, and helps establish the role of the Dpr and DIP in the formation of neuromuscular junctions.
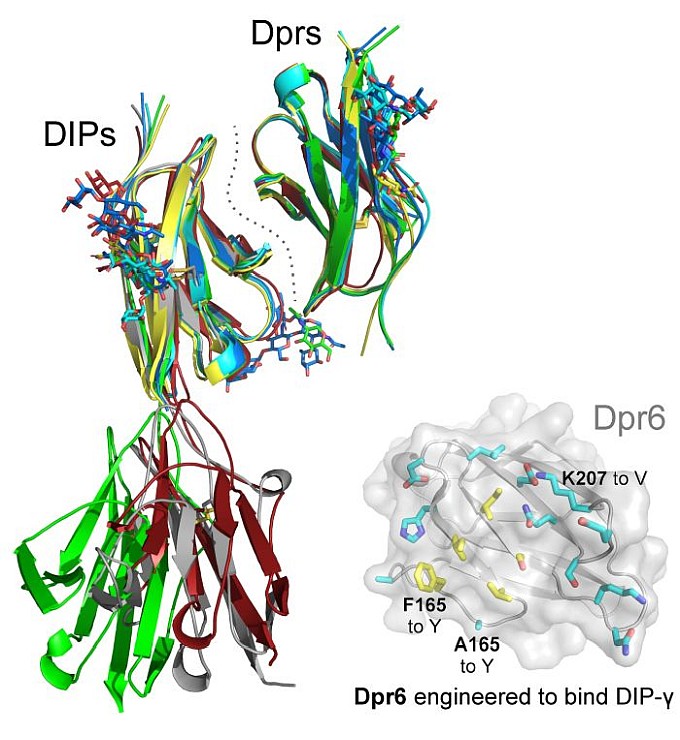 |
Figure: Overlaid structures of Dpr-DIP complexes, and mutational design for the engineering of Dpr6 to recognize DIP-gamma as a receptor. |
Citation: Cheng, S, Ashley, J, Kurleto, JD, Lobb-Rabe, M, Park, YJ, Carrillo, RA, Özkan, E., Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila, eLIFE J. 8, e41028-1-e41028-27 (2019). DOI: 10.7554/eLife.41028.001.