Led by Drs. Romila Mascarenhas and Markus Ruetz in Ruma Banerjee's laboratory at the University of Michigan, the group reported the crystal structure of human B12-dependent methylmalonyl-CoA mutase (MMUT) in complex with its G-protein chaperone, MMAA. This complex is central for supporting odd-chain fatty acid and branched amino acid catabolism, but its structure has been elusive due to its dynamic nature and variable stoichiometry. A clear image of the interacting proteins emerged from solution of the ring-shaped structure of the "M2C2" complex, comprising two MMUT and two MMAA dimers. Complex formation triggers large conformational changes; MMAA induces a dramatic 180-degree flipping out of the MMUT B12 binding domain while itself undergoing a 60-degree twist to complete its GTPase active site. These significant conformational reorientations help explain the detrimental effects of a subset of mutations at the newly formed MMUT-MMAA interfaces, which lead to the metabolic disease methylmalonic aciduria.
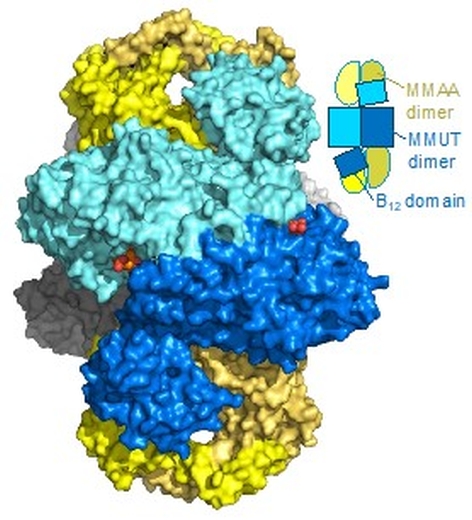 |
Figure: The M2C2 structure shown in surface and cartoon representation. The two MMUT dimers in the core are in blue and grey, while the two MMAA dimers at the ends are in yellow. The dark and light yellow shades represent each monomeric chains in the MMAA dimer. |
Citation: Mascarenhas, R, Ruetz, M, Gouda, H, Heitman, N, Yaw, M, Banerjee, R (2023) Architecture of the human G-protein-methylmalonyl-CoA mutase nanoassembly for B(12) delivery and repair. Nat Commun 14, 4332.